#include <som.h>
Inherits AbstractModel.
Inheritance diagram for SOM:

Public Types | |
| typedef std::list< std::pair< int, double > > | Neighbours |
| typedef std::vector< std::pair< matrix::Matrix, double > > | Neighbourhood |
Public Member Functions | |
| SOM () | |
| SOM (unsigned int dimensions, double sigma, double eps, double rbfsize, const std::string &name="SOM", const std::string &revision="$Id: som.h,v 1.5 2008/04/17 14:54:45 martius Exp $") | |
| create a som | |
| virtual | ~SOM () |
| virtual void | init (unsigned int inputDim, unsigned int outputDim, double unit_map=0.0, RandGen *randGen=0) |
| initialised som | |
| virtual const matrix::Matrix | process (const matrix::Matrix &input) |
| passive processing of the input (this function is not constant since a recurrent network for example might change internal states | |
| virtual const matrix::Matrix | learn (const matrix::Matrix &input, const matrix::Matrix &nom_output, double learnRateFactor=1) |
| virtual void | damp (double damping) |
| damps the weights and the biases by multiplying (1-damping) | |
| virtual unsigned int | getInputDim () const |
| returns the number of input neurons | |
| virtual unsigned int | getOutputDim () const |
| returns the number of output neurons | |
| virtual bool | store (FILE *f) const |
| stores the object to the given file stream (binary). | |
| virtual bool | restore (FILE *f) |
| loads the object from the given file stream (binary). | |
| virtual void | printWeights (FILE *f) const |
| const Neighbourhood & | getNeighbourhood () |
Public Attributes | |
| double | eps |
| learning rate for weight update | |
Protected Member Functions | |
| void | initNeighbourhood (double sigma) |
| initialised neighbourhood | |
| Neighbours | getNeighbours (int winner) |
| returns neighbourhood as a list of indices with weights | |
Static Protected Member Functions | |
| static double | activationfunction (void *rdfsize, double d) |
| activation function (rbf) | |
| static bool | validCoord (const matrix::Matrix &m, int size) |
| checks whether the given coordinate is within the lattice | |
| static int | coordToIndex (const matrix::Matrix &m, int size) |
| converts index to coordinates (size is the size of the space) | |
| static matrix::Matrix | indexToCoord (int index, int size, int dimensions) |
| converts coordinates to index (size is the size of the space) | |
Implementation follows normal vector-quantiser scheme. The output of the network is 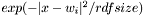 for each neuron.
for each neuron.
| typedef std::vector<std::pair<matrix::Matrix,double> > Neighbourhood |
| typedef std::list< std::pair<int,double> > Neighbours |
| SOM | ( | ) |
| SOM | ( | unsigned int | dimensions, | |
| double | sigma, | |||
| double | eps, | |||
| double | rbfsize, | |||
| const std::string & | name = "SOM", |
|||
| const std::string & | revision = "$Id: som.h,v 1.5 2008/04/17 14:54:45 martius Exp $" | |||
| ) |
create a som
| dimensions | number of dimensions of the neuron lattice |
| virtual ~SOM | ( | ) | [inline, virtual] |
| double activationfunction | ( | void * | rdfsize, | |
| double | d | |||
| ) | [static, protected] |
activation function (rbf)
| int coordToIndex | ( | const matrix::Matrix & | m, | |
| int | size | |||
| ) | [static, protected] |
converts index to coordinates (size is the size of the space)
| virtual void damp | ( | double | damping | ) | [inline, virtual] |
| virtual unsigned int getInputDim | ( | ) | const [inline, virtual] |
| const Neighbourhood& getNeighbourhood | ( | ) | [inline] |
| SOM::Neighbours getNeighbours | ( | int | winner | ) | [protected] |
returns neighbourhood as a list of indices with weights
| virtual unsigned int getOutputDim | ( | ) | const [inline, virtual] |
| Matrix indexToCoord | ( | int | index, | |
| int | size, | |||
| int | dimensions | |||
| ) | [static, protected] |
converts coordinates to index (size is the size of the space)
| void init | ( | unsigned int | inputDim, | |
| unsigned int | outputDim, | |||
| double | unit_map = 0.0, |
|||
| RandGen * | randGen = 0 | |||
| ) | [virtual] |
initialised som
| inputDim | dimension of input vector | |
| outputDim | number of outputneurons (must be a multiple of "dimensions" given at constructor) | |
| unit_map | if zero then weights are randomly choosed, otherwise uniformly distributed in the inputspace of size (unit_map x unit_map x ...) |
initialise neighbourhood
Implements AbstractModel.
| void initNeighbourhood | ( | double | sigma | ) | [protected] |
initialised neighbourhood
normalised gaussian
| const Matrix learn | ( | const matrix::Matrix & | input, | |
| const matrix::Matrix & | nom_output, | |||
| double | learnRateFactor = 1 | |||
| ) | [virtual] |
Implements AbstractModel.
| void printWeights | ( | FILE * | f | ) | const [virtual] |
| const Matrix process | ( | const matrix::Matrix & | input | ) | [virtual] |
passive processing of the input (this function is not constant since a recurrent network for example might change internal states
Implements AbstractModel.
| bool restore | ( | FILE * | f | ) | [virtual] |
| bool store | ( | FILE * | f | ) | const [virtual] |
| bool validCoord | ( | const matrix::Matrix & | m, | |
| int | size | |||
| ) | [static, protected] |
checks whether the given coordinate is within the lattice
| double eps |
learning rate for weight update
 1.4.7
1.4.7